Note
Go to the end to download the full example code.
Parallel Files#
The VTK library supports parallel file formats. Reading meshes broken up into several files is natively supported by VTK and PyVista.
from __future__ import annotations
import os
from pathlib import Path
import pyvista as pv
from pyvista import examples
Let’s go ahead and download the sample dataset containing an
pyvista.UnstructuredGrid broken up into several files.
Let’s inspect where this downloaded our dataset by setting load=False and
looking at the directory containing the file we downloaded.
['T0000000500.pvtu', 'T0000000500']
os.listdir(str(Path(path) / "T0000000500"))
['001.vtu', '003.vtu', '000.vtu', '002.vtu']
Note that a .pvtu file is available alongside a directory. This
directory contains all the parallel files or pieces that make the whole mesh.
We can simply read the .pvtu file and VTK will handle putting the mesh
together. In PyVista, this is accomplished through pyvista.read().
mesh = pv.read(filename)
mesh
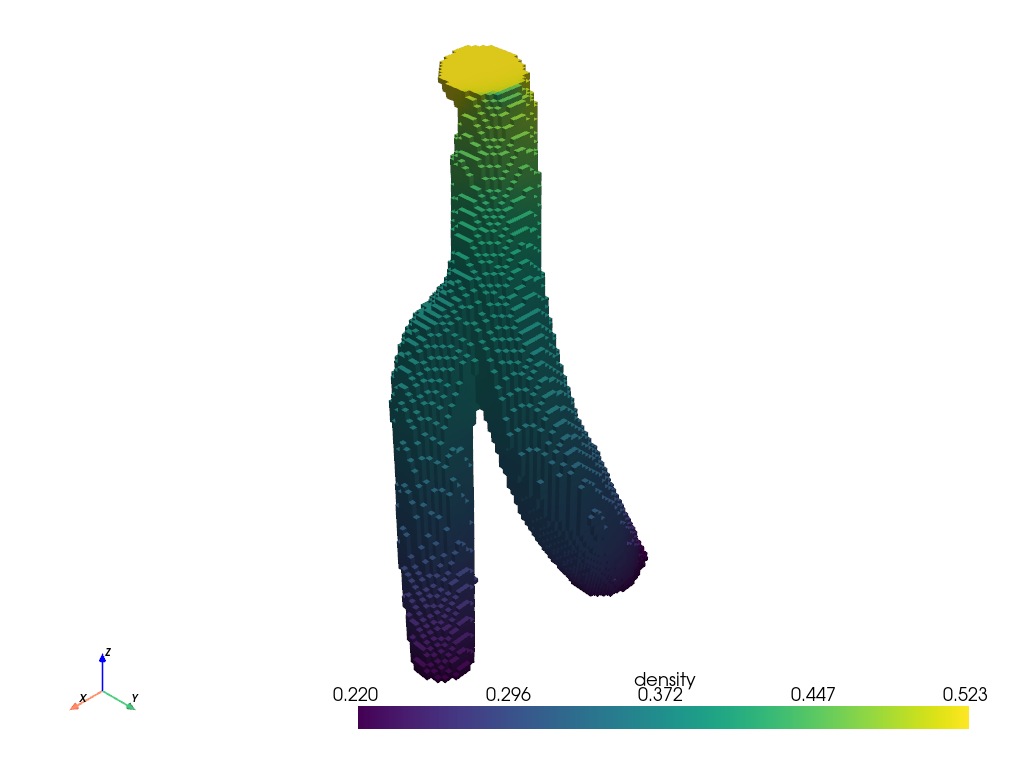
Plot the pieced together mesh
mesh.plot(scalars="node_value", categories=True)

mesh.plot(scalars="density")
Total running time of the script: (0 minutes 1.190 seconds)
