pyvista.examples.examples.load_frog_tissues#
- load_frog_tissues()[source]#
Load frog tissues dataset.
This dataset contains tissue segmentation labels for the frog dataset.
Added in version 0.44.0.
- Returns:
pyvista.ImageDataDataset.
Examples
Load data
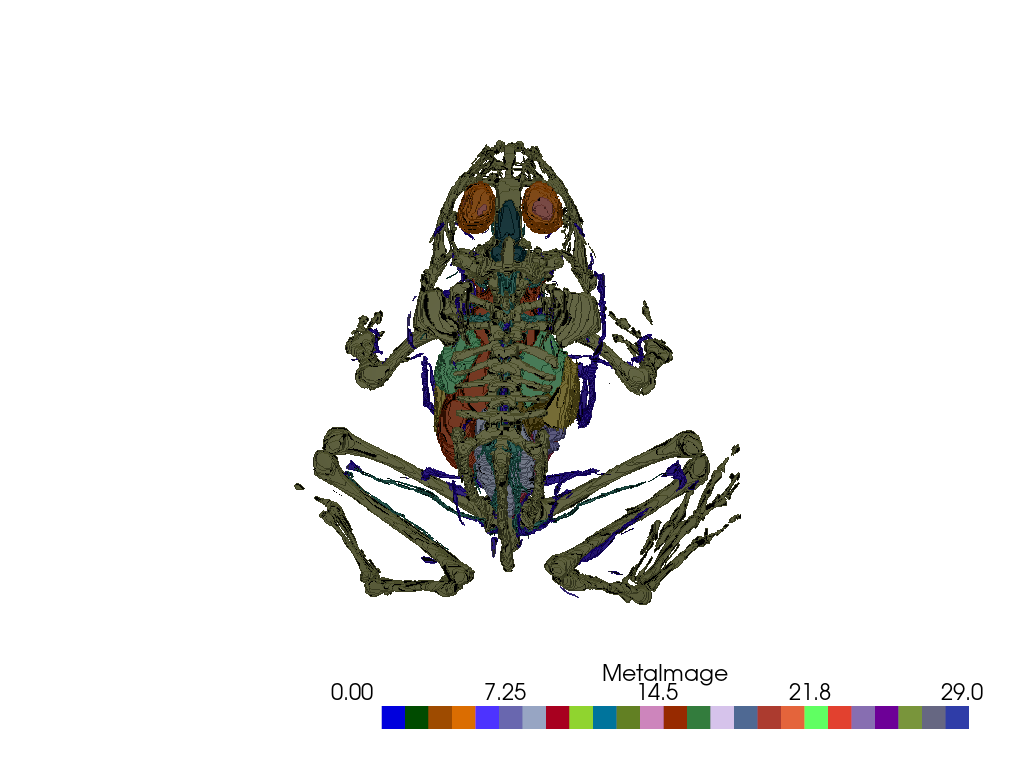
>>> import numpy as np >>> import pyvista as pv >>> from pyvista import examples >>> data = examples.load_frog_tissues()
Plot tissue labels as a volume
First, define plotting parameters
>>> # Configure colors / color bar >>> clim = data.get_data_range() # Set color bar limits to match data >>> cmap = 'glasbey' # Use a categorical colormap >>> categories = True # Ensure n_colors matches number of labels >>> opacity = 'foreground' # Make foreground opaque, background transparent >>> opacity_unit_distance = 1
Set plotting resolution to half the image’s spacing
>>> res = np.array(data.spacing) / 2
Define rendering parameters
>>> mapper = 'gpu' >>> shade = True >>> ambient = 0.3 >>> diffuse = 0.6 >>> specular = 0.5 >>> specular_power = 40
Make and show plot
>>> pl = pv.Plotter() >>> _ = pl.add_volume( ... data, ... clim=clim, ... ambient=ambient, ... shade=shade, ... diffuse=diffuse, ... specular=specular, ... specular_power=specular_power, ... mapper=mapper, ... opacity=opacity, ... opacity_unit_distance=opacity_unit_distance, ... categories=categories, ... cmap=cmap, ... resolution=res, ... ) >>> pl.camera_position = 'yx' # Set camera to provide a dorsal view >>> pl.show()
See also
- Frog Tissues Dataset
See this dataset in the Dataset Gallery for more info.
- Medical Datasets
Browse other medical datasets.