pyvista.examples.downloads.download_whole_body_ct_male#
- download_whole_body_ct_male(load=True, *, high_resolution=False)[source]#
Download a CT image of a male subject with 117 segmented anatomic structures.
This dataset is subject
's1397'from the TotalSegmentator dataset, version 2.0.1, available from zenodo. See the original paper for details:Jakob Wasserthal et al., TotalSegmentator: Robust Segmentation of 104 Anatomic Structures in CT Images, Radiology, Jul. 2023, doi: https://doi.org/10.1148/ryai.230024.
The dataset is loaded as a
MultiBlockwith three blocks:'ct':ImageDatawith CT data.'segmentations':MultiBlockwith 117ImageDatablocks, each containing a binary segmentation label. The blocks are named by their anatomic structure (e.g.'heart') and are sorted alphabetically. See the examples below for a complete list label names.'label_map':ImageDatawith a label map array. The label map is an alternative representation of the segmentation where the masks are combined into a single scalar array.Note
The label map is not part of the original data source.
Licensed under Creative Commons Attribution 4.0 International.
Added in version 0.45: Three dictionaries are now included with the dataset’s
user_dictto map label names to ids and colors:'names_to_colors': maps segment names to 8-bit RGB colors.'names_to_ids': maps segment names to integer ids used by the label map.'ids_to_colors': maps label ids to colors.
The label ids are the ids used by the included label map.
Changed in version 0.45: A downsampled version of this dataset with dimensions
(160, 160, 273)is now returned. Previously, a high-resolution version with dimensions(320, 320, 547)was returned. Usehigh_resolution=Truefor the high-resolution version.- Parameters:
- loadbool, default:
True Load the dataset after downloading it when
True. Set this toFalseand only the filename will be returned.- high_resolutionbool, default:
False Set this to
Trueto return a high-resolution version of this dataset. By default, aresampledversion with a0.5sampling rate is returned.Added in version 0.45.
- loadbool, default:
- Returns:
- output
pyvista.MultiBlockorstr DataSet or filename depending on
load.
- output
Examples
Load the dataset and get some of its properties.
>>> from pyvista import examples >>> import pyvista as pv >>> dataset = examples.download_whole_body_ct_male()
Get the CT image.
>>> ct_image = dataset['ct'] >>> ct_image ImageData (...) N Cells: 6876432 N Points: 6988800 X Bounds: 7.500e-01, 4.778e+02 Y Bounds: 7.500e-01, 4.778e+02 Z Bounds: 7.527e-01, 8.182e+02 Dimensions: 160, 160, 273 Spacing: 3.000e+00, 3.000e+00, 3.005e+00 N Arrays: 1
Get the segmentation label names and show the first three.
>>> segmentations = dataset['segmentations'] >>> label_names = segmentations.keys() >>> label_names[:3] ['adrenal_gland_left', 'adrenal_gland_right', 'aorta']
Get the label map and show its data range.
>>> label_map = dataset['label_map'] >>> label_map.get_data_range() (np.uint8(0), np.uint8(117))
Show the
'names_to_colors'dictionary with RGB colors for each segment.>>> dataset.user_dict['names_to_colors']
Show the
'names_to_ids'dictionary with a mapping from segment names to segment ids.>>> dataset.user_dict['names_to_ids']
Create a surface mesh of the segmentation labels.
>>> labels_mesh = label_map.contour_labels()
Color the surface using
color_labels(). Use the'ids_to_colors'dictionary that’s included with the dataset to map the colors.>>> colored_mesh = labels_mesh.color_labels( ... colors=dataset.user_dict['ids_to_colors'] ... )
Plot the CT image and segmentation labels together.
>>> pl = pv.Plotter() >>> _ = pl.add_volume( ... ct_image, ... cmap='bone', ... opacity='sigmoid_8', ... show_scalar_bar=False, ... ) >>> _ = pl.add_mesh(colored_mesh) >>> pl.view_zx() >>> pl.camera.up = (0, 0, 1) >>> pl.camera.zoom(1.3) >>> pl.show()
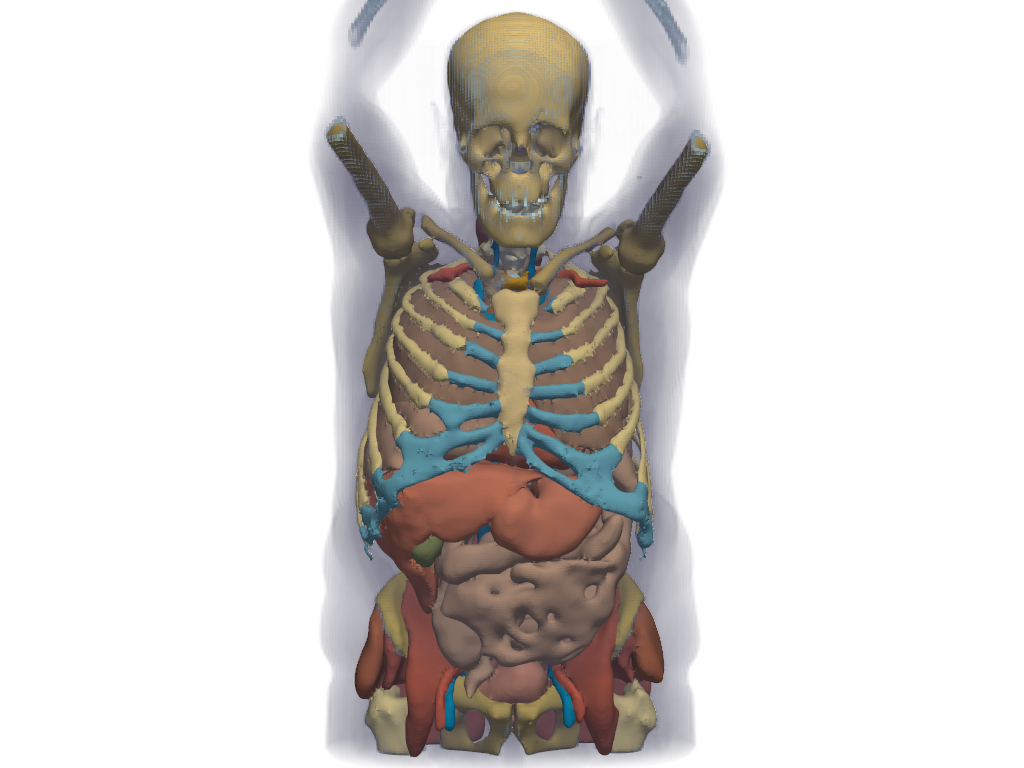
See also
- Visualize Anatomical Groups
Additional examples using this dataset.
- Whole Body Ct Male Dataset
See this dataset in the Dataset Gallery for more info.
- Whole Body Ct Female Dataset
Similar dataset of a female subject.
- Medical Datasets
Browse other medical datasets.
- Crop Labeled ImageData
Example cropping this dataset using a segmentation mask.
- Volume With Segmentation Mask
See additional examples using this dataset.